Girodat Group
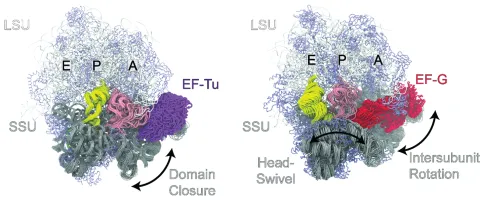
The Girodat Lab combines cutting-edge experimental and computational approaches to explore how the dynamic motions of ribonucleoproteins (RNPs) drive their biological functions. By integrating enzyme kinetics, advanced fluorescence techniques, molecular dynamics simulations, and cryo-electron microscopy, we aim to generate high-resolution, time-resolved insights into the structural dynamics of these essential molecular machines.
While traditional structural biology offers static snapshots, our work goes further—capturing the flexible, moving parts of RNPs that are crucial for their activity. This dynamic perspective provides a deeper understanding of how structure informs function, enabling discoveries that support both fundamental science and biomedical innovation.
At the core of our research is the ribosome—the universal RNP complex responsible for protein synthesis in all living cells. Current projects in the Girodat Lab include:
1. Engineering ribosome dynamics to control protein production by targeting key intersubunit bridges and hinge regions.
2. Deciphering how Ribosomal Protection Proteins alter ribosome dynamics to confer resistance to antibiotics.
3. Investigating disease-linked ribosomal mutations to understand how they disrupt normal dynamics and function.
Our work lays the foundation for developing next-generation antibiotics, improving therapeutic strategies, and optimizing protein synthesis in synthetic biology applications. Students in the Girodat Lab gain hands-on experience with interdisciplinary techniques and contribute to research at the forefront of molecular bioscience.
Student Positions
The Girodat lab currently has the following student positions available:
1. Structural Dynamics of Disease-Linked Ribosomes: Investigate how mutations in ribosomal RNA or proteins associated with human diseases alter ribosome structure and function, using integrative approaches such as cryo-electron microscopy, fluorescence spectroscopy, and molecular dynamics simulations.
2. Mechanisms of Antibiotic Resistance: Explore how bacterial ribosomes and associated proteins adapt to evade antibiotics, focusing on the structural dynamics of ribosomal protection proteins and their interactions with the ribosome.
3. Engineering Protein Synthesis and RNP Function: Design and test strategies to reprogram ribonucleoprotein dynamics for enhanced protein production or novel functions, integrating experimental and computational approaches in molecular biology, biophysics, and synthetic biology.
